The Mysterious Microbes That May Explain What Happened When Life Became Complex

An artist's impression of the relationships between the Asgard archea
Copyright American Institute of Physics (reprinting information)
(Inside Science) -- All life on Earth fits into three bins: Archaea, bacteria and eukaryotes. The first two bins are single-celled microbes, and the third includes animals, plants and fungi. Only the cells of the eurkaryotes contain the important elements known as nuclei, mitochondria, chloroplasts (in plants), and numerous others. The tough part is that nobody really knows exactly how or when life morphed from single-celled microbes into complex, multicellular eukaryotes.
The first life on Earth is thought to have begun as primitive cells forming in mineral pockets of primordial rocks nearly four billion years ago. Somewhat later came a Last Universal Common Ancestor, or LUCA, from which all subsequent life descended. Then, somewhere between 2.7 billion and 2.1 billion years ago the first traces of eukaryotes appear as telltale chemical signatures in rocks and later as actual fossils. For many evolutionary biologists, the details of that 600 million-year “moment” provoke burning curiosity and sometimes fiery debate. Did eukaryotes develop directly from some primordial mix of proto-life units? Or did they arise independently from the common ancestor or from the lineage of archaea? When and how did they manage to acquire the genes that code for the proteins that make it possible for them to construct their complex internal structure?
Read more of Inside Science's coverage of Earth's early life
Scientists Resurrect Ancient Versions of Life's Molecular Machinery
A Beautiful, Perplexing Ancient Crab
Fossil Bonanza in China Reveals Secrets from the Dawn of Animal Life
Loki the trickster
Until 1977, scientists thought there was only one kind of microbe -- bacteria -- but then microbiologist Carl Woese showed that archaea are a second kind, and that these two microbial domains diverged billions of years ago. We humans are closer to the archaea than to bacteria, but this is nearly the only undisputed statement among researchers who study how eukaryotes originated.
In 2015 a team of researchers at Uppsala University in Sweden led by Thijs Ettema announced they had found a group of archaea, dubbed Lokiarchaeota, that possess genes previously known only in the younger and more advanced eukaryotes. These microbes were discovered more than a mile below the ocean surface, in seafloor sediments near a hydrothermal feature on the floor of the Arctic Ocean, known as Loki’s Castle. Loki is the Norse trickster god.
When University of Texas microbiologist Brett Baker learned of the Lokiarchaeota and their eukaryotic genes, it was “totally shocking,” he said.
He had been sequencing the genome of a group of archaea found in sediment samples from North Carolina’s White Oak River and immediately realized his archaea must be closely related to the Loki group. “It completely changed my lab’s research,” he said of the Swedish discovery.
In 2016 Baker and colleagues published a paper naming his microbe Thorarchaeota after the Norse thunder god. Since then Baker has been collaborating with Thijs Ettema, the principal investigator of the Lokiarchaeota, who is now at the University of Wageningen in the Netherlands. Several related archaea, also named after Norse gods, have now been described, and the whole group is called the Asgard archaeota. (Asgard is the home of a tribe of Norse gods.)
When the Lokiarchaeota research was published, many press accounts called them the “missing link” between archaea and eukaryotes. Some scientists, led by microbiologist Patrick Forterre of the Pasteur Institute in Paris, strongly disagreed with the accuracy of the term and also with where Ettema placed eukaryotes within the archaeal branch of the tree of life. Forterre suggested that Ettema’s group used flawed theoretical reasoning and imprecise methods. Ettema’s group has fired back with similar attacks on Forterre’s methods.
Biology's event horizon
Organizing life forms according to the evolution of their genes and proteins is called phylogenomics. The genomics revolution of the last few decades transformed the field. It allows researchers to identify the smoking guns of ancestry and to trace the path of evolutionary bullets through time, revealing evolution in finer detail and through vastly more species than ever before. Capturing a modern genome tells scientists which proteins an organism could use. This gives scientists the chance to infer the character of ancient, lost life forms that lived in mud and mineral deposits eons ago.
However, to go back even further and understand the origins of the first eukaryotes, the field must wrestle with an apparent singularity in evolution known as endosymbiosis. Either by invasion or ingestion, a bacterium set up shop inside an archaeon, and that became essentially the first eukaryotic cell. The theory, advanced in 1967 by microbiologist Lynn Margulis, suggests that this accidental chocolate-and-peanut butter moment gave rise to modern eukaryotic cells, all of which possess mitochondria -- the remnants of the original bacterial symbiont.
Though this theory was only grudgingly accepted at first, it has become the dominant explanation for how eukaryotes emerged. Until the Asgard archaeota were found, endosymbiosis seemed to adequately explain the sudden emergence of this whole new type of cell. But the Asgard archaeota suggest that some of the genes for internal structures like nuclei, microtubules, Golgi bodies, and pores for molecules to pass through membranes may have been in the archaea before the eukaryotes emerged.
Mystery solved? Not entirely. Loki is a trickster, after all. Questions still abound. Since archaea -- including the Loki branch -- have no obvious internal subdivisions as eukaryotes do, what was the function of those eukaryotic genes? Just when did they appear? Scientists would dearly love to pin down not just what the Last Universal Common Ancestor was, but also the very first eukaryote and the last common ancestor of all modern eukaryotes. That would help scientists estimate the details of the emergence of the full eukaryotic toolkit and the point where eukaryotes began to differentiate from each other. But the timeline is still so murky that some scientists have described the path from first eukaryote to that last common ancestral eurkaryote as “effectively an event horizon,” evoking the mysterious edge of a black hole beyond which no light can escape.
At present, said Massachusetts Institute of Technology phylogeneticist Greg Fournier, “Eukaryogenesis represents a long protracted evolutionary event and there’s no one point where we can say, ‘Oh, here’s a eukaryote.’”
Global gene trade muddies the waters
Unfortunately, bringing that long, complicated process into better focus is subject to a major wild card: namely, horizontal gene transfer. Most modern eukaryotes reproduce by combining the genomes of their parents, in what’s called “vertical” transmission. But bacteria and archaea reproduce by processes like budding or cloning and maintain a lively global trade in genes as a separate process, transmitting them horizontally across species and even domains -- including to and from eukaryotes.
Horizontal gene transfer events can be difficult to track, and dating them can be extremely difficult. “Genes pop up from somewhere completely different,” said Peter Gogarten, a molecular biologist at the University of Connecticut at Storrs. There is also “a lot of turnover,” he added.
“After a gene transfer, you can see the recipient group, but we don’t know where along the branch that the recipient occurred,” said Fournier. “Oftentimes the genes have been lost in the donor group.”
Forterre said there were likely many types of early eukaryotes of which every trace has vanished from life’s record, and we can’t assume that modern archaea tell us much about them at all.
“All innovations had to take place during a long period of time,” Forterre said, “and many lineages predate the ancestors of modern eukaryotes.” Thus, while the Asgard archaea may be the closest living relatives of eukaryotes, they are probably not the group from which eukaryotes emerged. Forterre is among those scientists who believe there must have been a bottleneck in the eukaryotic lineage, when eukaryotes’ closest relatives died completely out, and only the eukaryotic branch survived.
Tracking genes through time
Genes and the proteins they encode tend to cluster into families. To determine evolutionary relationships, scientists first identify a genome or a set of genes and then analyze the proteins those genes specify. Some proteins are universally shared by all life, and researchers assume they are the oldest ones.
To trace the history of individual genes or proteins, scientists identify a group of related organisms with similar gene clusters, and then they see how those genes and the proteins they encode change over time. As cells reproduce, random changes to a cell's DNA result in slightly different protein chains, and over time different organisms will diverge. Scientists can compare these changes to trace how, and approximately when, the groups diverged from their common ancestor.
Horizontally transferred genes challenge the ability to do this, however, because both the original and transferred genes continue to evolve, and may gain novel functions in the original or the new genomic context, Fournier said.
Such issues create the kinds of disagreements that exist between the French and Dutch groups. Forterre's caustic critique alleged that what Ettema claimed to be eukaryotic material could have come from horizontal gene transfer. Forterre also claimed that the eukaryotic-specific proteins aren’t all present in all Asgard phyla, but are distributed in an uneven way, which supports the idea that they arrived horizontally, perhaps even from eukaryote to archaeon and not vice versa.
Even researchers who accept the Asgard data are quick to caution that the so-called eukaryotic genes found in the Asgard archaeota may not be acting like they would in a true eukaryote.
“It's completely inconceivable that they could have been sitting around without a function,” said Eugene Koonin, senior investigator at the National Center for Biotechnology Information, a component of the National Institutes of Health. But, he added, “functions change and it’s very common that in evolution preexisting parts are used in a new way. It’s entirely imaginable that this is what happened.”
Ettema agrees. “It will be interesting to see what they are doing in the Asgard archaeota,” he said.
Nesting conflicts
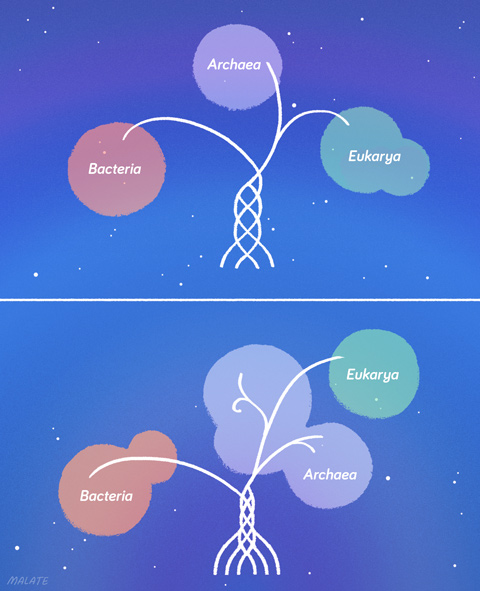
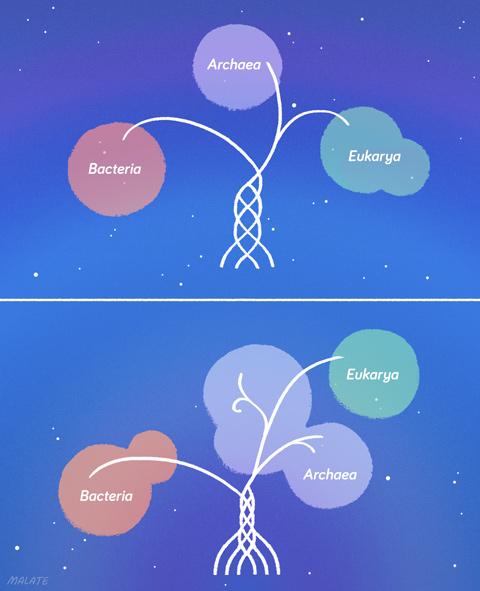
The Ettema-Forterre argument is nested inside a wider but equally contentious one involving many more scientists: the three-domain versus two-domain problem. The distinction between the three-domain and two-domain hypotheses is, to say the least, confusing.
Current conventional wisdom based on Woese’s 1970s argument is that LUCA branched into bacteria and archaea, and then archaea developed a sister branch, the eukaryotes. Most experts agree that eukaryotes are much closer to archaea than to bacteria, but just where and when eukaryotes became their own lineage is unsettled.
Think about humans and chimpanzees. We know we’re very closely related to chimpanzees, but we also know we’re not descended from them. Instead, our similarity stems from a common ancestor. But if we were descended from chimpanzees, we might consider the two species part of the same direct line.
If there are two domains -- bacteria and all other types of life -- then the last common ancestor of both archaea and eukaryotes might have had some proto-eukaryotic proteins, but others came into being in eukaryotes during the long period after archaea and eukaryotes diverged. This would explain why the Asgard archaeota have some but not all of those eukaryotic proteins. It also suggests that the endosymbiotic moment would be more likely to be the singular event that set eukaryotes forever apart from archaea. That event never happened in Asgard. It would keep eukaryotes in the archaeal line but as a very distinct lineage.
The domain argument has remained robust and unresolved for decades. But Ettema’s group supports the two-domain view.
“At some point our results started to change the basic tree of life from the three-domain to the two-domain,” he said, and there ensued a “fierce debate with several established microbiologists in the United States that didn’t agree with us, [whose] argument was based on historical grounds and personal views.”
His critics include some of the stellar experts in the field, like microbiologist Norman Pace at the University of Colorado Boulder, who thinks that the method Ettema used to compare gene segments, called concatenation, is unreliable.
Patrick Forterre takes issue with another part of Ettema’s approach. He said that by using Ettema’s method his team was able to produce either a two-domain or a three-domain tree depending on which protein they analyzed, when the result should not be dependent on the protein. “It's not a methodology problem,” he said. “It is a problem of interpretation.”
Scientific just-so stories
No matter how contrary to one another, all eukaryogenetic theories share two common problems: not enough information but a plethora of raw data and useful but flawed methods of analyzing it. The archaea have been dark and mysterious ever since Woese revealed their existence, and researchers are just beginning to parse the burgeoning data provided by genomics. This environment stimulates both fanciful and mundane conjectures. Accordingly, other scientists have offered flamboyant explanations of eukaryogenesis ranging from panspermia (they came from outer space) to viruses (which ferry genes into microbes and eukaryotes routinely) and variations of the hypotheses now under investigation. Many of these are what Gogarten calls “hand-waving” or “just-so stories,” because they have little or no supporting evidence. They can still serve a useful function, however, if they lead to fruitful investigations.
For example, he said, it would be interesting to know if there are other endosymbiotic organisms out there comprising an archaeon and a bacterium. So far, we have evidence suggestive of only one in all of evolution. Baker has seen hints of relationships between archaea and as yet unidentified bacteria indicating mutually beneficial metabolic processes in microbes living off hydrocarbons on or under the seafloor. “We know that there’s a genome out there very closely related to mitochondria that hasn’t been cultivated, and our next big goal is to figure out who that is,” said Baker.
But, cautioned Ettema, “the fact that you find interactions between archaea and bacteria doesn’t mean it’s something on its way to become a eukaryote. In nature there’s plenty of examples of archaea and bacteria that form some type of symbiosis” without merging into a single organism.
The role of technology and logistical cost/benefit ratios
For now most of what we know about the archaea comes from metagenomics -- grabbing samples from the environment and sorting the DNA by known chemistry and function, not by species. But that approach doesn’t reveal a whole organism, its physical properties or its actual cellular structure. So to really understand the Asgard archaeota, other approaches need to be applied.
In single cell genomics, the DNA of an individual microbe can be sequenced, vastly reducing the influence of contamination or processing artifacts. It became available only about a dozen years ago and is not yet universally practiced. Ettema’s team was unable to identify the Lokiarchaeota using single cell technology, so they turned to the metagenomic approach.
Another approach is to culture microbes in the lab in order to directly observe their properties in context. Microbiology has historically neglected archaea, in part because there are no confirmed examples of archaeal pathogens, and because they are notoriously difficult to grow in the lab. They tend to frequent dark, anoxic, high-temperature, acidic environments that are hard enough to sample, let alone duplicate from scratch.
“I think one reason we have so many difficulties in culturing these organisms is their own genome is barely enough to make a living,” Gogarten said. “In nature there are a lot of other organisms that provide services.” When a pure culture is attempted in the lab, it often fails for this reason. But new archaea have recently been found in less extreme environments around the world and these may be more amenable to lab life.
A tree with ziplines
The pursuit of eukaryotic origins and the vast evidence of gene trading across domains raises the problem of whether the longstanding metaphor of life as a tree makes sense any more.
“Certainly a tree does not describe major endosymbiotic events,” said Gogarten.
Single-celled organisms have never fit comfortably in the tree, whose structure is heavily skewed toward large eukaryotes -- not surprising considering eukaryotes devised the metaphor. But it’s now obvious that the tree must have a lot of ziplines winding horizontally through the canopy -- perhaps as many ziplines as branches.
In a sense, the ferment in the field means it’s right in the scientific sweet spot: big questions becoming clearer, and technologies emerging to answer them. This is a bit of a turnaround for microbiology. “When I started thinking about focusing my research [on archaea], people told me this would be scientific suicide,” Ettema said. “I wouldn’t have predicted this would lead to the revival of this whole field.”
Forterre’s group has praised the Ettema team, even while dismissing them, writing in a blog post that they did “fantastic work … building the new Asgard genomes and min[ing] them” even though he considers Ettema’s phylogenetic details to be inaccurate.
Thus, despite all the professional conflict, strange pieces of evidence and the near impossibility of finding an indisputable answer, microbiology has been energized by the Asgard archaeota. Valerian Dolja, a professor at Oregon State University’s Center for Genome Research and Biocomputing, calls the discovery of Asgard archaeota thrilling and "eye-opening.”
“Instead of just dreaming about what could have happened, we can compare everything to everything and figure out at least believable scenarios,” Dolja said.
The hope, he said, is that the effort will reduce the amount of hand-waving and tightly join together the many pieces of this complicated, tangled evolutionary story from the ancient and murky past.